Hey there,
I’m currently working on my Master’s thesis about federated learning and membership inference attacks. For my experiments, I deployed Flower using the Deployment Engine on our university HPC cluster (SLURM).
My setup consists of:
-
6 nodes total (1 server node + 5 client nodes)
-
Flower v1.22.0 (initially built with v1.14.0 and later upgraded)
-
Python 3.12 + Conda
-
fraction-fit = 1.0 (since I want to use all clients each round)
-
Each experiment runs about 10-40 min
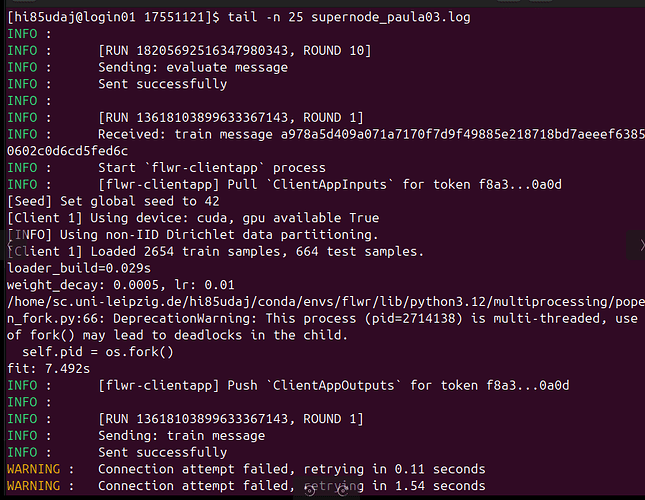
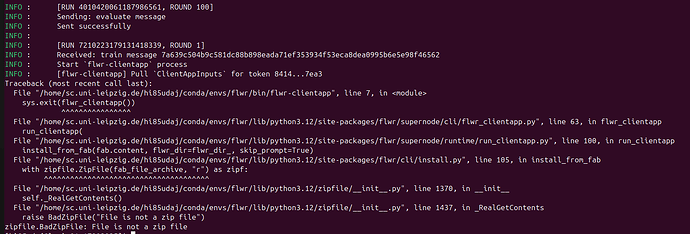
Everything works fine for several runs, but one client consistently loses connection to the server after about 7 hours of experiments. The previous experiment completes successfully (10 server rounds), but as soon as the next run starts, this client fails to reconnect to the server — resulting in a hang or timeout at server round 1 (screenshot).
From the logs, it looks like:
-
The previous run finishes properly.
-
The next run starts, but one client never re-establishes a connection.
Has anyone encountered similar issues when using Flower with long HPC jobs or multiple consecutive deployments via the Deployment Engine?
Could this be related to: Client processes not being fully cleaned up after a completed run?
Thanks in advance
#!/bin/bash
#SBATCH --job-name=flower-cluster
#SBATCH --output=logs/flower_%j_%N.out
#SBATCH --error=logs/flower_%j_%N.err
#SBATCH --partition=xx
#SBATCH --nodes=6
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=8
#SBATCH --mem=64G
#SBATCH --time=15:00:00
#SBATCH --gres=gpu:1
# --- Environment Setup ---
# --nodes=3 -> set client number(nodes-1) also in the experiment_config.flower.base.yaml
set -euo pipefail
module purge
mkdir -p logs
LOG_DIR="logs/${SLURM_JOB_ID}"
mkdir -p "$LOG_DIR"
echo "Logs will be saved in: $LOG_DIR"
# activate conda environment
module load Anaconda3/2024.02-1
eval "$(conda shell.bash hook)"
conda activate ~/conda/envs/flwr
# check python
which python
python -V
# check wandb login
python - <<'PY'
import wandb
print("wandb version:", wandb.__version__)
print("login ok:", wandb.login(relogin=False)) # uses saved key; returns True if configured
PY
# nvidia-smi # shows Driver Version and “CUDA Version”
# --- Distribute Flower ---
# Node 0 = Server, Nodes 1-9 = Clients
echo "SLURM_JOB_ID: $SLURM_JOB_ID"
echo "Running on nodes:"
scontrol show hostnames $SLURM_JOB_NODELIST
# --- Define Variables ---
PROJECT_DIR="$HOME/masterarbeit/Flower2"
NODES=($(scontrol show hostnames $SLURM_JOB_NODELIST))
SERVER_NODE=${NODES[0]}
CLIENTS=("${NODES[@]:1}")
NUM_CLIENTS=${#CLIENTS[@]}
CLIENT_HOSTS=$(IFS=, ; echo "${CLIENTS[*]}")
PORT_FLEET=9092 # SuperNodes -> SuperLink (Fleet API)
PORT_CTRL=9093 # flwr run -> SuperLink (CTRL API)
f
echo "Server node: $SERVER_NODE Fleet:${PORT_FLEET} CTRL:${PORT_CTRL}"
echo "Client nodes: ${CLIENTS[*]}"
echo "Project dir: $PROJECT_DIR"
# --- Prepare Data Splits (once per experiment) ---
SHARED_SPLITS_PATH="/work/flower/dataset_splits"
mkdir -p "$SHARED_SPLITS_PATH"
# Generate splits only if missing to fast hpc working dir
if [ ! -d "$SHARED_SPLITS_PATH/D1" ]; then
python - <<PY
from pathlib import Path
from flower.utils.split_cifar10_mia import split_cifar10_for_target_and_shadow
target = Path("$SHARED_SPLITS_PATH")
print("Writing CIFAR-10 splits to:", target)
split_cifar10_for_target_and_shadow(split_save_dir=target)
print("✅ Wrote splits to:", target)
PY
fi
# 1) Start SuperLink on server node (background)
srun --nodes=1 --nodelist="$SERVER_NODE" bash -lc "
cd '$PROJECT_DIR'
mkdir -p logs
flower-superlink \
--insecure \
--fleet-api-address 0.0.0.0:${PORT_FLEET} \
--control-api-address 0.0.0.0:${PORT_CTRL} \
--fleet-api-num-workers 8 \
> ${LOG_DIR}/superlink.log 2>&1
" &
echo "SuperLink started on ${SERVER_NODE}"
sleep 3 # give it time to bind
# 2) Start ALL SuperNodes in ONE srun (one task per client node)
# Each task gets a unique PARTITION_ID via SLURM_PROCID (0..NUM_CLIENTS-1)
srun --nodes="$NUM_CLIENTS" --ntasks="$NUM_CLIENTS" --ntasks-per-node=1 \
--nodelist="$CLIENT_HOSTS" --exclusive --kill-on-bad-exit=1 bash -lc "
cd '$PROJECT_DIR' && mkdir -p logs
PARTITION_ID=\$SLURM_PROCID
LOCAL_SPLITS_PATH=\"/dev/shm/cifar_splits\"
mkdir -p \"\$LOCAL_SPLITS_PATH\"
rsync -a --delete \"$SHARED_SPLITS_PATH/\" \"\$LOCAL_SPLITS_PATH/\"
export SPLITS_DIR=\"\$LOCAL_SPLITS_PATH\"
echo \"[Node \$SLURMD_NODENAME] Using SPLITS_DIR=\$SPLITS_DIR\"
flower-supernode \
--insecure \
--superlink ${SERVER_NODE}:${PORT_FLEET} \
--node-config \"partition-id=\${PARTITION_ID} num-partitions=${NUM_CLIENTS}\" \
> ${LOG_DIR}/supernode_\${SLURMD_NODENAME}.log 2>&1
" &
echo "SuperNodes started on client nodes: ${CLIENTS[*]}"
# 3) Wait until SuperLink Exec API is reachable
python - <<PY
import socket, time, sys
host="${SERVER_NODE}"; port=${PORT_CTRL}
for _ in range(180):
try:
with socket.create_connection((host, port), timeout=2):
print("SuperLink Exec API reachable at %s:%s" % (host, port)); sys.exit(0)
except OSError:
time.sleep(1)
print("ERROR: SuperLink Exec API NOT reachable"); sys.exit(1)
PY
# 4) Trigger the run from the server node (non-interactive)
SERVER_ADDRESS="${SERVER_NODE}:${PORT_CTRL}"
echo "🌼 Starting multiple Flower runs sequentially on $SERVER_NODE"
srun --nodes=1 --ntasks=1 --ntasks-per-node=1 --nodelist="$SERVER_NODE" --overlap bash -lc "
cd '$PROJECT_DIR'
# Inject correct server address into pyproject.toml
sed -i '/^\[tool\.flwr\.federations\.hpc-deploy\]/,/^\[/{
s/^address *= *.*/address = \"${SERVER_ADDRESS}\"/
}' pyproject.toml
# Sequentially run multiple Flower experiments
for run_id in \$(seq 6 35); do
echo \"🚀 Starting Flower run\${run_id}\"
python -u run_flower_hpc.py \"flower=main_mia_experiments/run\${run_id}\" 2>&1 | tee \"${LOG_DIR}/flwr_run\${run_id}.log\"
echo \"✅ Finished Flower run\${run_id}\"
echo
# optional small pause for W&B sync or disk I/O
sleep 3
done
# Revert .toml so it's clean for next job
sed -i '/^\[tool\.flwr\.federations\.hpc-deploy\]/,/^\[/{
s/^address *= *.*/address = \"SERVER_NODE:9093\"/
}' pyproject.toml
"
echo "✅ All Flower processes finished successfully."
Note: I am using Hydra to inject the experiment parameters to flower